Achieving reproducible results, whether working with full analytical pipelines or simple analyses, is contingent on the specific software used and the underlying environment. Often times, we need to reproduce experimental results or need to use packages that are no longer being maintained. With R, this is easily achieved through some combination of Conda virtual environments, renv and Docker images. This is often considered the epitome of reproducibility. However, sometimes it just doesn’t merit setting up a whole environment to work on a small project, therefore having multiple versions of R in the base environment might be desirable to speed things up. The process of setting up different R versions in the base environment, accessed through RStudio, depends on your operating system:
- Windows:
- Access RStudio’s Global Options to easily set your preferred binary version.
- Mac:
- You can install R in various ways with the most seamless method being Rswitch.
- Additional Resources:
- For installation details on Windows, Mac and Linux, check Posit’s support page.
- See Monica Thieu’s post for further insights from the community on managing multiple R versions on Mac.
How to install and switch between various versions of R in a Linux base environment?
It quite straightforward but like many tasks on Linux, it requires a bit more forethought and understanding of the system. Posit also has a good support page , but the seamless switching is only available in their Pro products. I decided to document how I did this on my local machine following Posit’s guides and the extensive documentation from the R Core Team. Along the way, I also opted to install R with the OpenBLAS library, which provides optimizedBLAS (Basic Linear Algebra Subprograms) and LAPACK (Linear Algebra PACKage) libraries to improve performance. An alternative BLAS/LAPACK library is the Intel oneAPI Base that is apparently faster. Perhaps I will try that later following instructions from Intel. For now, I’ll stick to OpenBLAS.
To provide a polished and comprehensive overview, I utilized ChatGPT, an AI language model by OpenAI, for drafting and refining portions of this article.
Let’s get into it
We’ll install R from source with two different configurations and learn how to switch between versions. See the R installation guide for more options.
Internal BLAS: Uses R’s internal shared BLAS library (
libRblas.so) for simplicity and modularity.OpenBLAS: Links R to system-optimized OpenBLAS and LAPACK libraries for performance gains.
How to switch between R versions: Changing environment variables to choose our R versions.
Both configurations include the shared R library (libR.so), which is important for compatibility with external applications like RStudio.
Configuration 2: Linking to System-Optimized OpenBLAS and LAPACK
Here, we want to link R to the system’s OpenBLAS and LAPACK libraries, which are optimized for performance and multi-threading. It is ideal for high-performance linear algebra operations. The way to achieve this is by explicitly requesting the libraries using the --with-blas and --with-lapack. Find more details in this old R installation guide and the upcoming R guide.
Prerequisites
Install the necessary build dependencies:
sudo pacman -S base-devel gcc-fortran openblas lapackThe main difference here is that we install openblas. It shouldn’t have any conflict with blas, if it does you might want to build from source in /opt as well.
Download and extract the R source code from CRAN:
wget https://cran.r-project.org/src/base/R-4/R-4.4.2.tar.gz
tar -xzvf R-4.4.2.tar.gz
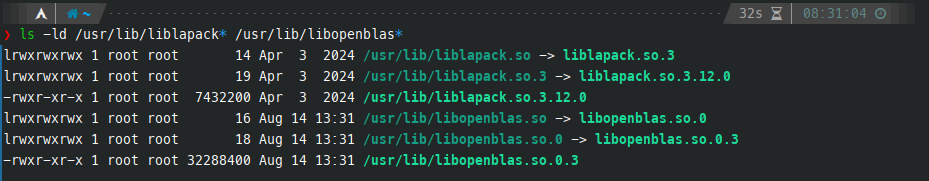
cd R-4.4.2We need to check where the BLAS and LAPACK library symlinks and executables are located:
ls -ld /usr/lib/liblapack* /usr/lib/libopenblas*I prefer to link the libraries by providing the absolute path. It is also possible to give a name without a path (like -lfoo) and the linker will look for a library file named libfoo.so (or libfoo.a for a static library) in standard library directories. See BLAS configuration guide.
blas-openblas and 64-Bit Integer Library Support
You can link to the bundled BLAS and LAPACK libraries provided by blas-openblas using the --with-blas flag.
Important: The blas-openblas library conflicts with blas. Since it provides BLAS/CBLAS/LAPACK/LAPACKE system-wide, any packages that depend on these libraries will need to be rebuilt. For this reason, building from source in a dedicated directory is recommended.
The blas64-openblas library does not create conflicts and can be particularly beneficial for handling large datasets or complex scientific computations. By supporting 64-bit indexing, blas64-openblas enables larger matrix operations, making it ideal for intensive data processing.
Installing R with openblas
Run the following command to configure R with OpenBLAS and LAPACK:
./configure --prefix=/opt/R/$(cat VERSION) \
--enable-R-shlib \
--with-blas="/usr/lib/libopenblas.so.0.3" \
--with-lapack="/usr/lib/liblapack.so.3.12.0"Flags used
--prefix=/opt/R/$(cat VERSION): Installs R in a versioned directory under/opt/R, as in Configuration 1.--enable-R-shlib: BuildslibR.so, the shared R library.--with-blas="/usr/lib/libopenblas.so.0.3": Links R to the system’s OpenBLAS library for optimized BLAS routines.--with-lapack="/usr/lib/liblapack.so.3.12.0": Links R to the system’s LAPACK library, providing optimized higher-level matrix operations.
Configuration output
R is now configured for x86_64-pc-linux-gnu
Source directory: .
Installation directory: /opt/R/4.4.2
C compiler: gcc -g -O2
Fortran fixed-form compiler: gfortran -g -O2
Default C++ compiler: g++ -std=gnu++17 -g -O2
C++11 compiler: g++ -std=gnu++11 -g -O2
C++14 compiler: g++ -std=gnu++14 -g -O2
C++17 compiler: g++ -std=gnu++17 -g -O2
C++20 compiler: g++ -std=gnu++20 -g -O2
C++23 compiler: g++ -std=gnu++23 -g -O2
Fortran free-form compiler: gfortran -g -O2
Obj-C compiler:
Interfaces supported: X11, tcltk
External libraries: pcre2, readline, BLAS(generic), LAPACK(generic), curl, libdeflate
Additional capabilities: PNG, JPEG, TIFF, NLS, cairo, ICU
Options enabled: shared R library, R profiling, libdeflate for lazyload
Capabilities skipped:
Options not enabled: shared BLAS, memory profiling
Recommended packages: yesNotice BLAS(generic), LAPACK(generic), indicating the use of the system’s libraries.
Building and installing R
Once configured with the above command, compile and install R:
make -j$(nproc)
sudo make installLike before, we run the following to check R’s configuration:
R sessionInfo()
Your output should look like this:
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Arch Linux
Matrix products: default
BLAS/LAPACK: /usr/lib/libopenblas.so.0.3; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
time zone: America/Chicago
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_4.4.Notice the linked openblas library in BLAS/LAPACK , in comparison to when we use --enable-BLAS-shlib only. OpenBLAS typically includes LAPACK routines as part of the library, so R is effectively using the LAPACK routines within OpenBLAS, which is compatible with LAPACK version 3.12.0.
Benefits of Using System-Optimized BLAS/LAPACK
By linking R to system-provided OpenBLAS and LAPACK libraries, we improved performance in numerical operations. Well, I haven’t performed a benchmark of my own but I trust that it is a little faster. We should expect:
- Performance Gains:
- OpenBLAS is highly optimized for operations such as matrix multiplications, dot products, and other foundational linear algebra tasks. It can significantly speed up computations in R, especially for functions that rely heavily on these operations (e.g., matrix decompositions, linear regression, and large-scale simulations).
- LAPACK complements BLAS by providing higher-level mathematical functions, such as eigenvalue decompositions, singular value decompositions (SVD), and other advanced matrix operations. LAPACK routines calls BLAS to perform complex computations, thus reducing computation time for common data science, statistical modeling, and machine learning operations.
- Multi-threading Support:
- OpenBLAS is designed to take advantage of multi-core CPUs, (with some caveats), making it ideal for computations that benefit from parallel processing. By linking R to OpenBLAS, R can automatically utilize multiple cores for matrix and linear algebra computations, significantly reducing computation times on multi-threaded operations.
- Architecture-Specific Optimizations:
- OpenBLAS and LAPACK are frequently optimized for specific CPU architectures (e.g., Intel, AMD) and can take advantage of processor-specific instructions like Advanced Vector Extensions (AVX) and Streaming SIMD Extensions (SSE). This means that R can achieve better performance on compatible hardware, albeit contingent on the underlying architecture, compared to using its internal BLAS/LAPACK libraries.
We could conduct benchmarks to measure how much computation time has improved, but that’s beyond our scope for now. To compare different R versions, I need to switch between them. So, how do we go about doing that?
How to switch between R versions
All that effort is impressive but it loses its value if it comes with friction. On Windows and Mac, toggling between R versions is straightforward, as I mentioned earlier. However, Linux users often face more challenges. Fortunately, the community has developed several solutions to streamline this process.
I found a nifty shim written by Jeff Keller to toggle between R versions. It’s a shell script to set the environment variable R_VERSION to the preferred version and then launch R Studio.
I echo his script here with some modifications of my own.
Toggle Shim
As a root, replace the R and Rscript binaries found in /bin/R/ and /bin/Rscript.
cat << 'EOF' /bin/R
#!/bin/bash
<<prelude
Shim from https://jeff.vtkellers.com/posts/data-science/managing-multiple-r-installations-on-linux/
used to toggle between R versions installed in /opt/R
prelude
# Uncomment this to set a fixed default version of R. Otherwise, the
# most recent version will be used.
R_VERSION_DEFAULT="4.4.1"
# The default installation directory is /opt/R (https://docs.rstudio.com/resources/install-r/)
# Change this as is necessary for your environment.
R_INSTALL_DIR="/opt/R"
R_VERSIONS_AVAIL=($(basename --multiple $(dirname $(dirname $(ls "${R_INSTALL_DIR}"/*/bin/R))) | sort --version-sort))
# If no default R version is set then set the most recent version as the default
if [ -z ${R_VERSION_DEFAULT} ]; then
R_VERSION_DEFAULT="${R_VERSIONS_AVAIL[-1]}"
fi
# If a specific R version was not requested then use the default
if [ -z ${R_VERSION} ]; then
R_VERSION="${R_VERSION_DEFAULT}"
fi
R_BINARY_BASE=$(basename $0)
R_BINARY="${R_INSTALL_DIR}/${R_VERSION}/bin/${R_BINARY_BASE}"
if [ ! -f $R_BINARY ]; then
echo "${R_BINARY_BASE} ${R_VERSION} not found. Available versions: ${R_VERSIONS_AVAIL[*]}"
exit 1
fi
# Unset the R_HOME env var if something else set it
unset R_HOME
"${R_BINARY}" "$@"
EOFChanging /bin/R/ should result in changes in /usr/bin/R. You can double check before proceeding. I fixed my default version of R but you can comment to set to the latest R version.
Make the script executable:
chmod +x /bin/RCreate a Symlink to Rscript
ln -s /bin/R /bin/RscriptUsage
export R_VERSION=4.4.1
rstudio &Conclusions
We’ve installed different versions of R and learned how to switch between them. This flexibility can help us experiment with different analyses and projects. The toggling shim is an excellent solution for launching RStudio with your preferred R version. However, I encountered an issue when opening R projects (.Rproj files). Regardless of the R version specified for the project, it always loads the latest installed version of R. Additionally, setting the R_VERSION environment variable does not affect which R version the project uses. This means that even if your project was created with an different version of R, it will still open with an unintended latest version in /opt/R.
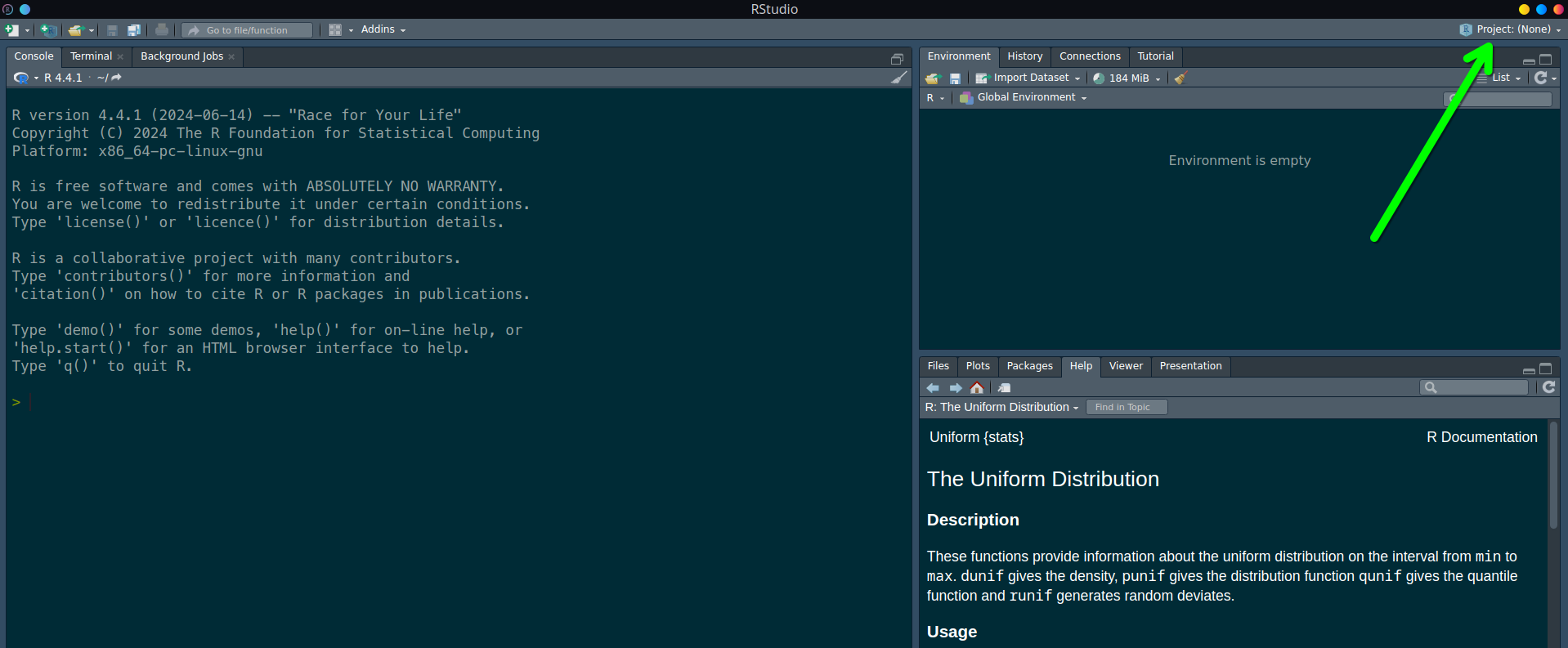
A practical workaround I found is to launch RStudio with the desired R version first, then switch to the intended project using the toggle in the upper right corner. See green arrow below.
It’s not a frictionless solution but it’s not too bad once you’ve incorporated it in your workflow. For now, it will do. I’ll slowly make improvements to see if I can get it working with projects.
I hope this helps out folks wanting to have various versions of R and easily switch between them. Please let me know if you have any comments of suggestions.
Additional Resources
Official Documentation
Installing Multiple Versions of R on Linux
Comprehensive guide from Posit on how to install and manage multiple R versions on Linux systems.Changing R Versions for the RStudio Desktop IDE
Instructions on how to switch between different R versions within the RStudio IDE.R Administration Manual
Official R documentation detailing configurations.
Community Discussions
R with LAPACK from OpenBLAS - Posit Forum
A forum thread discussing the integration of LAPACK from OpenBLAS in R.Faster BLAS in R - Brett Klamer’s Blog
Blog post exploring methods to speed up BLAS operations in R.
Tutorials and Guides
R Performance with BLAS - csantill.github.io
A detailed tutorial on optimizing R performance using BLAS libraries.Why is R Slow? Explanations and MKL/OpenBLAS Setup to Fix It
An R-bloggers article explaining common performance issues in R and how to address them with MKL/OpenBLAS.Monica Thieu’s Post on Managing Multiple R Versions
Insights and solutions from the community on handling multiple R versions.
Tools and Repositories
- r-blas GitHub Repository
GitHub repository for managing BLAS configurations in R.